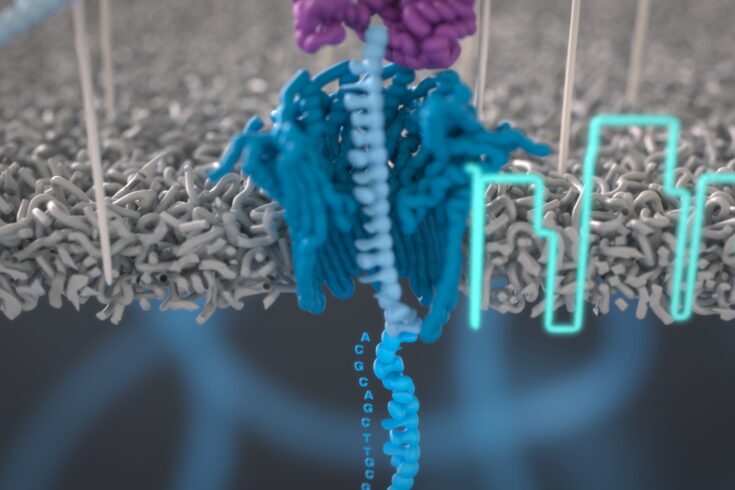
Nearly 25 years on from the first human genome being sequenced, whole genome sequencing has scaled up substantially. Whole genome sequencing means determining the order of the 6.4 billion DNA building blocks in an individual’s genetic code.
The technologies developed have led to large-scale sequencing. This includes a project that’s under way to sequence the whole genomes of half a million people, participants of UK Biobank. We look back at four key milestones in our history that have helped pave the way, and the impact of genomics on people’s lives today.
1. Discovering the structure of DNA
Credit: Medical Research Council.
Video transcript and on-screen captions are available by watching on YouTube.
In the 1940s, the Medical Research Council (MRC) set up two new research units. Based in Cambridge and London these aimed to study the structure of biological molecules. This early investment into the newly developing field of structural biology paid off. Scientists Dr James Watson and Professor Francis Crick joined the MRC Molecular Structure of Biological Systems Unit. Meanwhile Professor Maurice Wilkins and Dr Rosalind Franklin began work at the MRC Biophysics Research Unit.
Together, the work of these four scientists led to the discovery of the now famous double helix structure of DNA. Watson and Crick won the Nobel Prize in 1962 for the discovery. The structure led to insights and new technologies that changed the landscape of cellular biology. It heralded a golden age of MRC-funded researchers looking at the potential uses of X-ray crystallography. The developing field of looking inside proteins to see their inner structures was born.
2. Developing DNA sequencing

Fred Sanger. Credit: MRC Laboratory of Molecular Biology
Fast forward to 1977, at the MRC Laboratory of Molecular Biology. There Dr Frederick Sanger developed a way to work out the exact sequences of bases in a DNA molecule. He used this new technique to decipher the genetic sequence of a virus, the first fully-sequenced genome. His method for sequencing DNA, known as the ‘Sanger sequencing method’ was key to the Human Genome Project. This led to the sequencing of the first human genome which increased the understanding of many genetic diseases and cancer. His work won him a Nobel Prize in 1980.
3. Next generation sequencing for cheaper, faster genome data
Credit: Oxford Nanopore Technologies
Video transcript and on-screen captions are available by watching on YouTube.
Discovery science in the 1980s led to the development of next generation sequencing. This technology performs automated sequencing, running many reactions at the same time. Parallel sequencing like this offers ultra-high throughput, scalability and speed. It made it possible to sequence a whole human genome at a fraction of the cost and time of traditional Sanger sequencing techniques.
One method is nanopore sequencing, which enables real-time analysis of long DNA or RNA fragments. Nanopore sequencing works by monitoring the changes to an electrical current as the DNA or RNA molecule passes through a tiny protein pore, called a nanopore. This innovation led to the founding of a spin-out company called Oxford Nanopore Technologies in 2005. The development of nanopore sequencing was supported by funding from multiple research councils, part of UK Research and Innovation.
4. Portable genome sequencing enabling bedside testing

Jaqueline Goes de Jesus and Nuno Faria use the Oxford Nanopore MinION device in front of the minibus equipped with lab in João Pessoa, Brazil. Credit: Richardo Funari.
Over recent years, advances in technology mean that clinicians can do diagnostic tests at patient bedsides rather than at the laboratory bench. Known as ‘point-of-care testing’, these technologies benefit both the patient and healthcare professionals. Quicker results mean faster treatment and less cost to the health service.
In the past, routine point-of-care testing was limited to biochemical tests such as measuring glucose or blood cell counts. But in the last decade, point-of-care diagnostic development has expanded. It’s now used in the fields of infectious diseases and personalised medicine, thanks to the ‘MinION sequencer’. Developed by Oxford Nanopore Technologies, this is a tiny pocket-sized DNA sequencer. The sequencer can perform nanopore-based sequencing in a few hours. It has helped track the spread of Ebola and Zika viruses, identify Tuberculosis pathogens in clinical samples and track key mutations in patient tumour samples.
Where sequencing goes from here
Large-scale genomic data offers the potential for scaling up from people to populations. And studying population genomics is key to understanding how human health and disease risk changes as we grow older.
The exciting area of ‘functional genomics’ looks at how differences in our genomes influence diseases. We’re supporting this through a major new functional genomics initiative. This research provides new information to increase our understanding of disease. And this knowledge is crucial for developing new diagnosis methods and potential therapies. Like identifying a drug that kills off bowel cancer cells that contain a specific gene mutation. This avoids the unpleasant side effects of traditional chemotherapy which kills both healthy and cancer cells. Development of new and quicker diagnosis methods will ensure people with treatable illnesses receive effective treatment faster.
Another promising area is studying human and non-human genomes, including pathogens. Every individual’s response to infection is different. Understanding these differences and identifying treatments according to the genetic make-up of pathogens, can help researchers develop more precise ways to tackle infections. For example:
- a UK-wide consortium showed how the hepatitis C virus uses mutations to hide from patients’ immune systems. They’ve trialled two new drugs, providing hundreds of patients with early access to these drugs, saving the NHS around £12 million
- a study looked at genetic differences in patients with COVID-19. Those with specific gene mutations had a higher risk of mild infection progressing into a life-threatening condition. The results highlighted which drugs should be prioritised for clinical testing
Decades of discovery science have fuelled a genomic revolution. It’s changing the future of healthcare and is set to continue improving health and wellbeing for patients.
Find out more
Explore our timeline of MRC research and discoveries to find out more about MRC-funded achievements.
Find further information on how we support genomics research.
UK Biobank is a vast biomedical database helping scientists and researchers to discover the triggers for different health outcomes along our lifecourse. MRC has funded UK Biobank from the beginning, along with the Wellcome Trust, the British Heart Foundation, Cancer Research UK and the National Institute of Health and Care Research.
Top image: DNA strand being fed through a nanopore and corresponding signal. Credit: Oxford Nanopore Technologies